About

Evolutionary Biologist
There is grandeur in this view of life...having been originally breathed into a few forms or into one; and that...from so simple a beginning endless forms most beautiful and most wonderful have been, and are being, evolved.
I am an evolutionary biologist (among other things) interested in understanding the processes that shape biodiversity. Most of my research has been conducted in snakes, with a special emphasis on understanding the molecular and evolutionary processes shaping the venom system in vipers. I conducted my PhD work in Dr. Christopher L. Parkinson's lab at the University of Central Florida and Clemson University. I am currently working as a postdoctoral research fellow with Dr. H. Lisle Gibbs at The Ohio State University. This website is still under construction.
Research
Phylogenomics & Reticulate Evolution
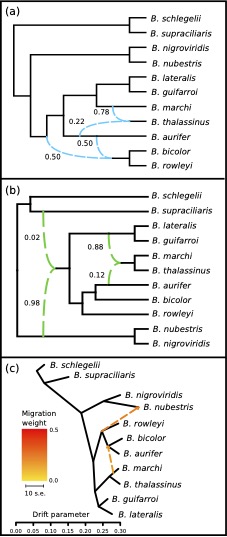
Massively parallel sequencing technologies have revolutionized efforts to resolve the tree of life by leveraging hundreds or thousands of genetic markers for phylogenetic reconstruction.
Simultaneously, we have gained a greater understanding of the processes generating phylogenetic patterns such as discordance or disagreement among different markers.
Processes like introgression and hybrid speciation are increasingly recognized for their important roles shaping evolutionary histories.
I am interested in untangling these often complex relationships and understanding their implications for the evolution of other biological traits
In a recent example, my collaborators and I studied the apparent phylogenetic discordance in palm-pitvipers using Anchored Phylogenomics.
This approach allowed us to sample hundreds of nuclear loci, and demonstrate key differences in the relationships inferred from nuclear and mitochondrial phylogenies.
To investigate what could be responsible for these different signals, we tested for evidence of reticulate evolution.
We found that multiple approaches suggested introgression of palm-pitviper lineages, and that these divergent signals corresponded to conflicting phylogenetic topologies.
You can read more about this study and access the original work here.
Deciphering Trait Evolution

Striking biological phenotypes are among the most iconic biological images, but in many cases our understanding of how adaptive traits emerge and are shaped by selection or drift is incomplete.
I am interested in understanding how biological traits emerge and diversify across species groups to further our knowledge of how phenotypic diversity is generated.
Snake venoms are an especially fruitful area of study because of the particularly strong and readily quantifyable relationship between genotype and phenotype.
Venoms are also remarkably variable at several biological levels, which provides an ideal setting for testing how differences emerge.
Some of my work to-date has correlated known or observed differences in venom phenotypes with particular genetic mechanisms to infer possible mechanisms of divergence.
Most recently I collaborated on a project examining how different sources of genetic variation, gene content, transcript sequence diversity, and expression diversity are associated with more or less phylogenetically diverse diets.
Combining phylogenetic path modeling with a recently published venom gland transcriptome dataset, we find that higher toxin sequence diversity is associated with broader diets.
Surpringly, higher expression diversity was associated with less broad diets, but with toxin family-specific analyses we found that this pattern was driven by PLA2s.
Unlike other large toxin families, overall higher PLA2 expression, rather than more diverse PLA2 expression, was associated with broader diets.
While linking specific sources of venom variation to distinct ecological outcomes, this study also demonstrated how distinct genomic regions can respond to selection differently.
Identifying Genomic Mechanisms of Differentiation
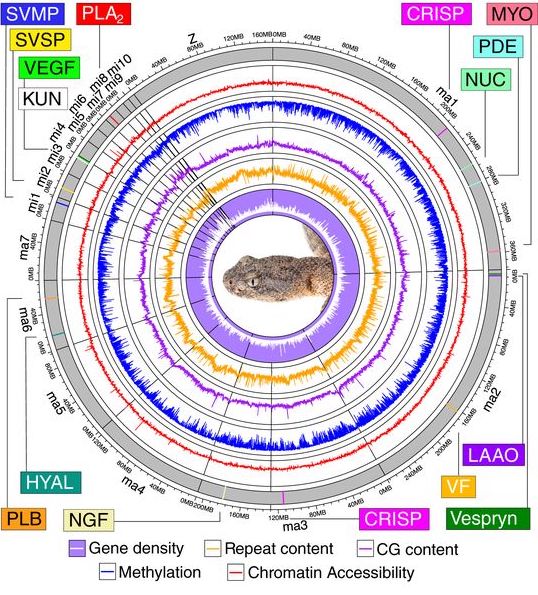
Differences that we observe among populations and species start with changes that occur at the genomic level. A wide array of genetic mechanisms can produce novel or modified phenotypes, but how various mechanisms contribute to phenotypic diversification independently or in concert is not well-understood in most cases. Using new sequencing technologies and emerging analytical methods, we can evaluate the roles of different genetic mechanisms in phenotypic differentiation. My current work in this topic has been primarily focused on identifying the genomic changes that are associated with different venom compositions in Sistrurus and other rattlesnakes.
Population Genomics of Founding Events
Population expansions into new habitats create opportunities for lineage divergence and may be associated with early stages of speciation. Founding events can occur naturally as species ranges expand or be human-mediated, as is the case in biological invasions. Such cases offer the opportunity to examine how population sizes and genetic diversity change during population establishment, and subsequently shape evolutionary tragectories. We are currently evaluating these topics in multiple systems, including mainlaind-island populations of Brazilian jararaca lineages and an invasive population of common wall lizards in Cinncinnatii, Ohio.
Publications
In Revision
Mason, A. J., Vasquez, C. R., Townsend, J. H., Herrera-B., L. A., Borja, M., Sasa, M., Parkinson, C. L. Family Matters: Comparative Phylogenetic Approaches Reveal Rapid Evolution and Adaptive Shifts in Expression in Diverse Venom Gene Families. Submitted to Systematic Biology. Decision: Reject; Resubmission encouraged.
2022
1. Holding, M. L., Trevine, V. C., Zinenko, O., Strickland, J. L., Rautsaw, R. M., Mason, A. J., Hogan, M. P., Parkinson, C. L., Grazziotin, F. G., Santanta, S. E., Davis, M. A.,and Rokyta, D. R. Evolutionary allometry and ecological correlates of fang length evolution in vipers.. Proceedings of the Royal Society B: Biological Sciences, 289(1982), 20221132.
2. Myers, E. A., Strickland, J. L., Rautsaw, R. M., Mason, A. J., Schramer, T. D., Nystrom, G. S., Hogan, M. P., Yooseph, S., Rokyta, D. R., & Parkinson, C. L. De novo genome assembly highlights the role of lineage-specific duplications in the evolution of venom in Fea’s Viper. Genome Biology and Evolution, 14(7), evac082.
Mason, A. J., Holding, M. H., Rautsaw, R. M., Rokyta, D. R., Parkinson, C. L., Gibbs, H. L. Venom gene sequence diversity and expression jointly shape diet adaptation in pitvipers. Molecular Biology & Evolution, 39(4), msac082.
Nachtigall, P. G., Freitas-de-Sousa, L. A., Mason, A. J., Moura-da-Silva, A. M., Grazziotin, F. G., & Junqueira-de-Azevedo, I. L. Differences in PLA2 constitution distinguish the venom of two endemic Brazilian mountain lanceheads, Bothrops cotiara and Bothrops fonsecai. Toxins, 14(4), 237.
2021
Nachtigall, P. G., Rautsaw, R. M. Ellsworth, S., Mason, A. J., Rokyta, D. R., Parkinson, C. L., Junqueira-de-Azevedo, I. L. M. ToxCodAn: A new toxin annotator and guide to venom gland transcriptomics. Briefings in Bioinformatics.
Holding, M. L., Stickland, J. L., Rautsaw, R. M.. Hofmann, E. P., Mason, A. J., M., Nystrom, G., Ellsworth, S. A., Colston, T. J., Borja, M., Castañeda-Gaytán, G., Grunwald, C. I., Jones, J. M., Freitas-de-Sousa, L., Viala, V. L., Margres, M. J., Zaher, E. H., Junqueira-de-Azevedo, I. L. M., Moura-de-Silva, A. M., Grazziotin, F. G., Gibbs, H. L., Rokyta, D. R., Parkinson, C. L. Phylogenetically diverse diets favor more complex venoms in North American pitvipers. Proceedings of the National Academy of Sciences. 118(17).
Hofmann, E. P., Rautsaw, R. M., Mason, A. J., Strickland, J. L., Parkinson, C.L Duvernoy’s Gland Transcriptomics of the Plains Black-HeadedSnake, Tantilla nigriceps (Squamata, Colubridae): Unearthing the Venom of Small Rear-Fanged Snakes. Toxins. 13(5), 336.
Margres, M. J., Rautsaw, R. M., Strickland, J. L., Mason, A. J., Schramer, T., Hofmann, Stiers, E., Bartlett, D., Colston, T.J., Rokyta, D. R., Parkinson, C. L. The Tiger Rattlesnake genome reveals how a complex genotype produces the simplest venom phenotype. Proceedings of the National Academy of Sciences. 118(4).
2020
Rautsaw, R. M., Schramer, T. D., Acuña, R., Arick, L. N., DiMeo, M., Mercier, K. P., Schrum, M., Mason, A. J., Margres, M. J., Strickland, J. L., Parkinson, C. L. Genomic adaptations to salinity resist gene flow in the evolution of Floridian Watersnakes. Molecular Biology and Evolution. 38(3),745-760.
Mason, A. J., Margres, M. J., Strickland, J. L., Sasa, M., Rokyta, D. R., Parkinson, C. L. Trait differentiation and modular toxin expression in Palm-Pitvipers. BMC Genomics, 21(1), 1-20.
2019
Rautsaw, R. M., Hofmann, E. P., Margres, M. J., Holding, M. L., Strickland, J. L., Mason, A. J., Rokyta, D. R., Parkinson, C. L. Intraspecific sequence variation and gene expression contribute little to venom diversity in the Sidewinder Rattlesnake (Crotalus cerastes). Proceedings of the Royal Society B: Biological Sciences. 286(1906), 20190810.
Mason, A. J., Grazziotin, R. A., Zaher, H., Lemmon, A. R., Lemmon, E. M., and Parkinson, C. L. Reticulate evolution in Nuclear Middle America causes discordance in the phylogeny of palm-pitvipers. Journal of Biogeography, 46(5), 833-844.
2018
Strickland, J. L., Smith, C. F., Mason, A.J., Borja, M., Castañeda-Gaytán, G., Schield, D.R., Castoe, T. A., Spencer, C. L., Smith, L. L., Trápaga, A., Bouzid, N. M., Campillo-García, G., Flores-Villela, O. A., Antonio-Rangel, D., Rokyta, D. R., Mackessy, S. P., and Parkinson, C. L. Evidence for divergent patterns of local selection driving venom variation in Mojave Rattlesnakes (Crotalus scutulatus). Scientific Reports, 8(1), 17622.
Hofmann, E. P., Rautsaw, R. M., Strickland, J. L., Holding, M. L., Hogan, M. P., Mason, A. J., Rokyta, D. R., and Parkinson, C. L. Comparative venom-gland transcriptomics and venom proteomics of four Sidewinder Rattlesnake (Crotalus cerastes) lineages reveal little differential expression despite individual variation. Scientific Reports, 8(1), 15534.
Holding, M., Margres, M. J., Mason, A. J., Parkinson, C. L., and Rokyta, D.R. Evaluating the performance of de novo assembly methods for venom-gland transcriptomics. Toxins, 10(6), 249.
Strickland, J. L., Mason, A. J., Rokyta, D. R., and Parkinson, C. L. Phenotypic Variation in Mojave Rattlesnake (Crotalus scutulatus) venom is driven by four toxin families. Toxins, 10(4), 135.
Whittington, C. A, Mason, A. J., and Rokyta, D.R. A single mutation unlocks cascading exaptations in the origin of a potent pitviper neurotoxin. Molecular Biology & Evolution. 35(4), 887-898.
2016
Doan, T. M., Mason, A.J., Castoe, T. A., Sasa, M. M., and Parkinson, C. L. A cryptic palm-pitviper species (Squamata: Viperidae: Bothriechis) from the Costa Rican highlands, with notes on the variation within B. nigroviridis. Zootaxa, 4138(2), 271-290.
Teaching
In addition to my focus on research, I enjoy teaching when the opportunity arises. This includes contributing to some of my collaborators courses like Advanced Evolution and Herpetology with guest lectures, lesson plans, and activities. Most recently, I have partnered with Dr. Eric Gangloff at Ohio Wesleyan University to build a teaching module on venom in herpetology. This unit covers a variety of evolutionary concepts such as identifying evidence of selection, inferring evolutionary patterns such as convergence, and understanding the genetic mechanisms that facilitate evolution. Material for this unit has been curated as a CUBES module here.
Outreach
Much like teaching, I have not had as many recent opportunities for outreach, but I really enjoy drawing on my time working at an AZA accredited zoo facility to convey scientific information to diverse audiences. While working on my PhD I was able to participate in a number of events such as Clemson University's Tiger Field Day, where we presented information on snakes to as many as 16,000 attendees. Recently, I have also worked with a student at Ohio Wesleyan Univeristy, Jaquelyn Kessar, to help put together an exhibit on snake venom for the university's natural history museum. Speaking for myself, I was super impressed with all the work Jaquelyn put in and I am really excited by how it turned out!


